有条件地将数据点指向R中的置信带之外
我需要为下面的图中的置信区域之外的数据点着色不同于带内的数据点。我应该在数据集中添加单独的列来记录数据点是否在置信带内吗?你能举个例子吗?
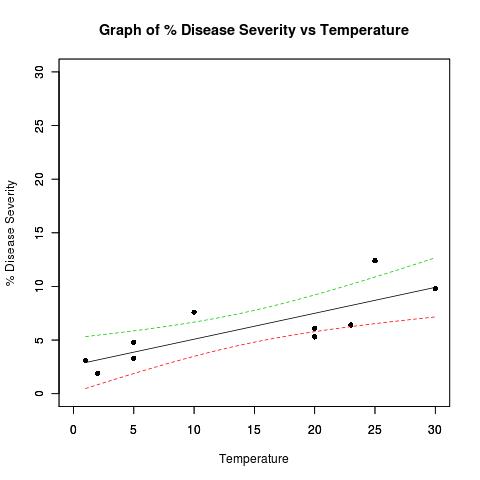
示例数据集:
## Dataset from http://www.apsnet.org/education/advancedplantpath/topics/RModules/doc1/04_Linear_regression.html
## Disease severity as a function of temperature
# Response variable, disease severity
diseasesev<-c(1.9,3.1,3.3,4.8,5.3,6.1,6.4,7.6,9.8,12.4)
# Predictor variable, (Centigrade)
temperature<-c(2,1,5,5,20,20,23,10,30,25)
## For convenience, the data may be formatted into a dataframe
severity <- as.data.frame(cbind(diseasesev,temperature))
## Fit a linear model for the data and summarize the output from function lm()
severity.lm <- lm(diseasesev~temperature,data=severity)
# Take a look at the data
plot(
diseasesev~temperature,
data=severity,
xlab="Temperature",
ylab="% Disease Severity",
pch=16,
pty="s",
xlim=c(0,30),
ylim=c(0,30)
)
title(main="Graph of % Disease Severity vs Temperature")
par(new=TRUE) # don't start a new plot
## Get datapoints predicted by best fit line and confidence bands
## at every 0.01 interval
xRange=data.frame(temperature=seq(min(temperature),max(temperature),0.01))
pred4plot <- predict(
lm(diseasesev~temperature),
xRange,
level=0.95,
interval="confidence"
)
## Plot lines derrived from best fit line and confidence band datapoints
matplot(
xRange,
pred4plot,
lty=c(1,2,2), #vector of line types and widths
type="l", #type of plot for each column of y
xlim=c(0,30),
ylim=c(0,30),
xlab="",
ylab=""
)
3 个答案:
答案 0 :(得分:10)
好吧,我认为使用ggplot2会很容易,但现在我意识到我不知道如何计算stat_smooth / geom_smooth的置信限制。
请考虑以下事项:
library(ggplot2)
pred <- as.data.frame(predict(severity.lm,level=0.95,interval="confidence"))
dat <- data.frame(diseasesev,temperature,
in_interval = diseasesev <=pred$upr & diseasesev >=pred$lwr ,pred)
ggplot(dat,aes(y=diseasesev,x=temperature)) +
stat_smooth(method='lm') + geom_point(aes(colour=in_interval)) +
geom_line(aes(y=lwr),colour=I('red')) + geom_line(aes(y=upr),colour=I('red'))
这会产生: alt text http://ifellows.ucsd.edu/pmwiki/uploads/Main/strangeplot.jpg
我不明白为什么stat_smooth计算的置信带与直接从predict(即红线)计算的带不一致。任何人都可以对此有所了解吗?
编辑:
想通了。 ggplot2使用1.96 *标准误差来绘制所有平滑方法的间隔。
pred <- as.data.frame(predict(severity.lm,se.fit=TRUE,
level=0.95,interval="confidence"))
dat <- data.frame(diseasesev,temperature,
in_interval = diseasesev <=pred$fit.upr & diseasesev >=pred$fit.lwr ,pred)
ggplot(dat,aes(y=diseasesev,x=temperature)) +
stat_smooth(method='lm') +
geom_point(aes(colour=in_interval)) +
geom_line(aes(y=fit.lwr),colour=I('red')) +
geom_line(aes(y=fit.upr),colour=I('red')) +
geom_line(aes(y=fit.fit-1.96*se.fit),colour=I('green')) +
geom_line(aes(y=fit.fit+1.96*se.fit),colour=I('green'))
答案 1 :(得分:6)
最简单的方法可能是计算TRUE/FALSE值的向量,以指示数据点是否在置信区间内。我将稍微重新调整您的示例,以便在执行绘图命令之前完成所有计算 - 这在程序逻辑中提供了一个清晰的分离,如果您将其中的一部分打包成一个函数,则可以利用它
第一部分基本相同,只是我用lm()变量替换predict()内的severity.lm附加调用 - 不需要使用额外的计算资源来重新计算当我们已经存储它时的线性模型:
## Dataset from
# apsnet.org/education/advancedplantpath/topics/
# RModules/doc1/04_Linear_regression.html
## Disease severity as a function of temperature
# Response variable, disease severity
diseasesev<-c(1.9,3.1,3.3,4.8,5.3,6.1,6.4,7.6,9.8,12.4)
# Predictor variable, (Centigrade)
temperature<-c(2,1,5,5,20,20,23,10,30,25)
## For convenience, the data may be formatted into a dataframe
severity <- as.data.frame(cbind(diseasesev,temperature))
## Fit a linear model for the data and summarize the output from function lm()
severity.lm <- lm(diseasesev~temperature,data=severity)
## Get datapoints predicted by best fit line and confidence bands
## at every 0.01 interval
xRange=data.frame(temperature=seq(min(temperature),max(temperature),0.01))
pred4plot <- predict(
severity.lm,
xRange,
level=0.95,
interval="confidence"
)
现在,我们将计算原始数据点的置信区间并运行测试以查看这些点是否在区间内:
modelConfInt <- predict(
severity.lm,
level = 0.95,
interval = "confidence"
)
insideInterval <- modelConfInt[,'lwr'] < severity[['diseasesev']] &
severity[['diseasesev']] < modelConfInt[,'upr']
然后我们将绘制情节 - 首先是高级绘图函数plot(),正如您在示例中使用的那样,但我们只绘制区间内的点。然后我们将跟进低级函数points(),它将以不同的颜色绘制区间外的所有点。最后,matplot()将用于填写您使用它时的置信区间。但是,我不是调用par(new=TRUE)而是将参数add=TRUE传递给高级函数,以使它们像低级函数一样。
使用par(new=TRUE)就好像在玩一个诡计多端的绘图功能 - 这可能会产生无法预料的后果。 add参数由许多函数提供,以使它们向绘图添加信息而不是重绘它 - 我建议尽可能利用这个参数,并作为最后的手段重新使用par()操作。 / p>
# Take a look at the data- those points inside the interval
plot(
diseasesev~temperature,
data=severity[ insideInterval,],
xlab="Temperature",
ylab="% Disease Severity",
pch=16,
pty="s",
xlim=c(0,30),
ylim=c(0,30)
)
title(main="Graph of % Disease Severity vs Temperature")
# Add points outside the interval, color differently
points(
diseasesev~temperature,
pch = 16,
col = 'red',
data = severity[ !insideInterval,]
)
# Add regression line and confidence intervals
matplot(
xRange,
pred4plot,
lty=c(1,2,2), #vector of line types and widths
type="l", #type of plot for each column of y
add = TRUE
)
答案 2 :(得分:4)
我喜欢这个主意,并试图为此制作一个功能。当然,它远非完美。欢迎您的意见
diseasesev<-c(1.9,3.1,3.3,4.8,5.3,6.1,6.4,7.6,9.8,12.4)
# Predictor variable, (Centigrade)
temperature<-c(2,1,5,5,20,20,23,10,30,25)
## For convenience, the data may be formatted into a dataframe
severity <- as.data.frame(cbind(diseasesev,temperature))
## Fit a linear model for the data and summarize the output from function lm()
severity.lm <- lm(diseasesev~temperature,data=severity)
# Function to plot the linear regression and overlay the confidence intervals
ci.lines<-function(model,conf= .95 ,interval = "confidence"){
x <- model[[12]][[2]]
y <- model[[12]][[1]]
xm<-mean(x)
n<-length(x)
ssx<- sum((x - mean(x))^2)
s.t<- qt(1-(1-conf)/2,(n-2))
xv<-seq(min(x),max(x),(max(x) - min(x))/100)
yv<- coef(model)[1]+coef(model)[2]*xv
se <- switch(interval,
confidence = summary(model)[[6]] * sqrt(1/n+(xv-xm)^2/ssx),
prediction = summary(model)[[6]] * sqrt(1+1/n+(xv-xm)^2/ssx)
)
# summary(model)[[6]] = 'sigma'
ci<-s.t*se
uyv<-yv+ci
lyv<-yv-ci
limits1 <- min(c(x,y))
limits2 <- max(c(x,y))
predictions <- predict(model, level = conf, interval = interval)
insideCI <- predictions[,'lwr'] < y & y < predictions[,'upr']
x_name <- rownames(attr(model[[11]],"factors"))[2]
y_name <- rownames(attr(model[[11]],"factors"))[1]
plot(x[insideCI],y[insideCI],
pch=16,pty="s",xlim=c(limits1,limits2),ylim=c(limits1,limits2),
xlab=x_name,
ylab=y_name,
main=paste("Graph of ", y_name, " vs ", x_name,sep=""))
abline(model)
points(x[!insideCI],y[!insideCI], pch = 16, col = 'red')
lines(xv,uyv,lty=2,col=3)
lines(xv,lyv,lty=2,col=3)
}
像这样使用:
ci.lines(severity.lm, conf= .95 , interval = "confidence")
ci.lines(severity.lm, conf= .85 , interval = "prediction")
- 我写了这段代码,但我无法理解我的错误
- 我无法从一个代码实例的列表中删除 None 值,但我可以在另一个实例中。为什么它适用于一个细分市场而不适用于另一个细分市场?
- 是否有可能使 loadstring 不可能等于打印?卢阿
- java中的random.expovariate()
- Appscript 通过会议在 Google 日历中发送电子邮件和创建活动
- 为什么我的 Onclick 箭头功能在 React 中不起作用?
- 在此代码中是否有使用“this”的替代方法?
- 在 SQL Server 和 PostgreSQL 上查询,我如何从第一个表获得第二个表的可视化
- 每千个数字得到
- 更新了城市边界 KML 文件的来源?