Matlab:seqlogo具有统一的绘图列高度
在Matlab中,我想制作seqlogo plot氨基酸序列谱。但是不是通过熵来缩放绘图列的高度,而是希望所有列都具有相同的高度。
我正在修改this question的答案中的代码,但我想知道是否有一个seqlogo参数或我错过的其他一些函数可以使列高度统一。< / p>
或者,是否有可以应用于序列配置文件的统计变换来破解所需的输出? (柱高均匀,每个字母的高度线性比例 它在seqprofile中的概率)
2 个答案:
答案 0 :(得分:2)
解决此问题的最简单方法可能是直接修改Bioinformatics Toolbox function SEQLOGO的代码(如果可能)。在R2010b中,您可以:
edit seqlogo
该功能的代码将显示在编辑器中。接下来,找到以下行(第267-284行),并将它们注释掉或完全删除它们:
S_before = log2(nSymbols);
freqM(freqM == 0) = 1; % log2(1) = 0
% The uncertainty after the input at each position
S_after = -sum(log2(freqM).*freqM, 1);
if corrError
% The number of sequences correction factor
e_corr = (nSymbols -1)/(2* log(2) * numSeq);
R = S_before - (S_after + e_corr);
else
R = S_before - S_after;
end
nPos = (endPos - startPos) + 1;
for i =1:nPos
wtM(:, i) = wtM(:, i) * R(i);
end
然后将这一行放在他们的位置:
wtM = bsxfun(@times,wtM,log2(nSymbols)./sum(wtM));
您可能希望以新名称保存文件,例如seqlogo_norm.m,因此您仍然可以使用原始未经修改的SEQLOGO功能。现在,您可以创建序列配置文件图,其中所有列标准化为相同的高度。例如:
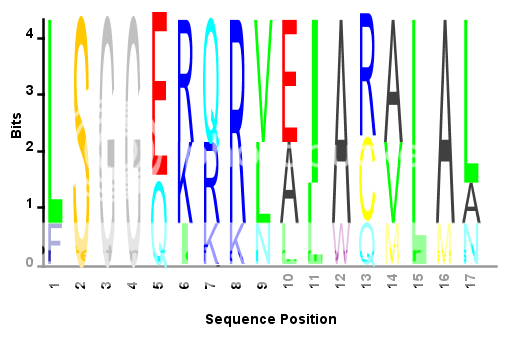
S = {'LSGGQRQRVAIARALAL',... %# Sample amino acid sequence
'LSGGEKQRVAIARALMN',...
'LSGGQIQRVLLARALAA',...
'LSGGERRRLEIACVLAL',...
'FSGGEKKKNELWQMLAL',...
'LSGGERRRLEIACVLAL'};
seqlogo_norm(S,'alphabet','aa'); %# Use the modified SEQLOGO function
OLD ANSWER:
我不确定如何转换序列配置文件信息以从Bioinformatics Toolbox function SEQLOGO获得所需的输出,但我可以向您展示如何修改我为{{3}编写的替代seqlogo_new.m你链接到。如果您更改初始化bitValues的行:
bitValues = W{2};
到此:
bitValues = bsxfun(@rdivide,W{2},sum(W{2}));
然后你应该将每列缩放到1的高度。例如:
S = {'ATTATAGCAAACTA',... %# Sample sequence
'AACATGCCAAAGTA',...
'ATCATGCAAAAGGA'};
seqlogo_new(S); %# After applying the above modification

答案 1 :(得分:1)
目前,我的解决方法是生成一堆与序列配置文件匹配的假序列,然后将这些序列提供给http://weblogo.berkeley.edu/logo.cgi。以下是制作假序列的代码:
function flatFakeSeqsFromPwm(pwm, letterOrder, nSeqsToGen, outFilename)
%translates a pwm into a bunch of fake seqs with the same probabilities
%for use with http://weblogo.berkeley.edu/
%pwm should be a 4xn or a 20xn position weight matrix. Each col must sum to 1
%letterOrder = e.g. 'ARNDCQEGHILKMFPSTWYV' for my data
%nSeqsToGen should be >= the # of pixels tall you plan to make your chart
[height windowWidth] = size(pwm);
assert(height == length(letterOrder));
assert(isequal(abs(1-sum(pwm)) < 1.0e-10, ones(1, windowWidth))); %assert all cols of pwm sum to 1.0
fd = fopen(outFilename, 'w');
for i = 0:nSeqsToGen-1
for seqPos = 1:windowWidth
acc = 0; %accumulator
idx = 0;
while i/nSeqsToGen >= acc
idx = idx + 1;
acc = acc + pwm(idx, seqPos);
end
fprintf(fd, '%s', letterOrder(idx));
end
fprintf(fd, '\n');
end
fclose(fd);
end
相关问题
最新问题
- 我写了这段代码,但我无法理解我的错误
- 我无法从一个代码实例的列表中删除 None 值,但我可以在另一个实例中。为什么它适用于一个细分市场而不适用于另一个细分市场?
- 是否有可能使 loadstring 不可能等于打印?卢阿
- java中的random.expovariate()
- Appscript 通过会议在 Google 日历中发送电子邮件和创建活动
- 为什么我的 Onclick 箭头功能在 React 中不起作用?
- 在此代码中是否有使用“this”的替代方法?
- 在 SQL Server 和 PostgreSQL 上查询,我如何从第一个表获得第二个表的可视化
- 每千个数字得到
- 更新了城市边界 KML 文件的来源?