如何从字符中非常有效地提取特定模式?
我有这样的大数据:
> Data[1:7,1]
[1] mature=hsa-miR-5087|mir_Family=-|Gene=OR4F5
[2] mature=hsa-miR-26a-1-3p|mir_Family=mir-26|Gene=OR4F9
[3] mature=hsa-miR-448|mir_Family=mir-448|Gene=OR4F5
[4] mature=hsa-miR-659-3p|mir_Family=-|Gene=OR4F5
[5] mature=hsa-miR-5197-3p|mir_Family=-|Gene=OR4F5
[6] mature=hsa-miR-5093|mir_Family=-|Gene=OR4F5
[7] mature=hsa-miR-650|mir_Family=mir-650|Gene=OR4F5
我想要做的是,在每一行中,我想在单词成熟= 之后选择名称,并在 Gene = 之后选择单词,然后再对它们进行描述与
一起paste(a,b, sep="-")
hsa-miR-5087-OR4F5
hsa-miR-26a-1-3p-OR4F9
所以,最终的实现是这样的:
for(i in 1:nrow(Data)){
Data[i,3] <- sub("mature=([^|]*).*Gene=(.*)", "\\1-\\2", Data[i,1])
Name <- strsplit(as.vector(Data[i,2]),"\\|")[[1]][2]
Data[i,4] <- as.numeric(sub("pvalue=","",Name))
print(i)
}
效果很好,但速度很慢。 数据的大小非常大,并且 200,000,000 行。这个实现非常缓慢。我怎样才能加快速度?
5 个答案:
答案 0 :(得分:11)
如果您可以保证格式与您指定的完全一致,那么正则表达式可以捕获(由下面的括号表示)从等号到管道符号,从Gene =到结尾的所有内容,然后粘贴它们与减号一起使用:
sub("mature=([^|]*).*Gene=(.*)", "\\1-\\2", Data[,1])
答案 1 :(得分:5)
另一种选择是将read.table与=一起用作分隔符,然后粘贴2列:
res = read.table(text=txt,sep='=')
paste(sub('[|].*','',res$V2), ## get rid from last part here
sub('^ +| +$','',res$V4),sep='-') ## remove extra spaces
[1] "hsa-miR-5087-OR4F5" "hsa-miR-26a-1-3p-OR4F9" "hsa-miR-448-OR4F5" "hsa-miR-659-3p-OR4F5"
[5] "hsa-miR-5197-3p-OR4F5" "hsa-miR-5093-OR4F5" "hsa-miR-650-OR4F5"
答案 2 :(得分:5)
已经给出的简单sub解决方案看起来相当不错,但以下是其他一些方法:
1)read.pattern 在gsubfn package中使用read.pattern,我们可以将数据解析为data.frame。然后可以通过多种方式操纵这种中间形式DF。在这种情况下,我们使用paste的方式与问题基本相同:
library(gsubfn)
DF <- read.pattern(text = Data[, 1], pattern = "(\\w+)=([^|]*)")
paste(DF$V2, DF$V6, sep = "-")
,并提供:
[1] "hsa-miR-5087-OR4F5" "hsa-miR-26a-1-3p-OR4F9" "hsa-miR-448-OR4F5"
[4] "hsa-miR-659-3p-OR4F5" "hsa-miR-5197-3p-OR4F5" "hsa-miR-5093-OR4F5"
[7] "hsa-miR-650-OR4F5"
生成的中间数据框DF如下所示:
> DF
V1 V2 V3 V4 V5 V6
1 mature hsa-miR-5087 mir_Family - Gene OR4F5
2 mature hsa-miR-26a-1-3p mir_Family mir-26 Gene OR4F9
3 mature hsa-miR-448 mir_Family mir-448 Gene OR4F5
4 mature hsa-miR-659-3p mir_Family - Gene OR4F5
5 mature hsa-miR-5197-3p mir_Family - Gene OR4F5
6 mature hsa-miR-5093 mir_Family - Gene OR4F5
7 mature hsa-miR-650 mir_Family mir-650 Gene OR4F5
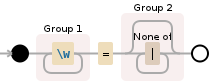
以下是我们使用的正则表达式的可视化:
(\w+)=([^|]*)
1a)名称我们可以通过分别读取三列数据和三个名称来使DF更好看。这也改进了paste语句:
DF <- read.pattern(text = Data[, 1], pattern = "=([^|]*)")
names(DF) <- unlist(read.pattern(text = Data[1,1], pattern = "(\\w+)=", as.is = TRUE))
paste(DF$mature, DF$Gene, sep = "-") # same answer as above
本节中生成的DF看起来像这样。它有3列而不是6列,其余列用于确定适当的列名称:
> DF
mature mir_Family Gene
1 hsa-miR-5087 - OR4F5
2 hsa-miR-26a-1-3p mir-26 OR4F9
3 hsa-miR-448 mir-448 OR4F5
4 hsa-miR-659-3p - OR4F5
5 hsa-miR-5197-3p - OR4F5
6 hsa-miR-5093 - OR4F5
7 hsa-miR-650 mir-650 OR4F5
2)strapplyc
使用相同包的另一种方法。这将提取a = a之后的字段,而不包含|制作一份清单。然后,我们将该第一和第三个字段粘贴在一起:
sapply(strapplyc(Data[, 1], "=([^|]*)"), function(x) paste(x[1], x[3], sep = "-"))
给出相同的结果。
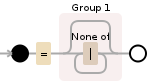
以下是使用的正则表达式的可视化:
=([^|]*)
答案 3 :(得分:4)
这是一种方法:
Data <- readLines(n = 7)
mature=hsa-miR-5087|mir_Family=-|Gene=OR4F5
mature=hsa-miR-26a-1-3p|mir_Family=mir-26|Gene=OR4F9
mature=hsa-miR-448|mir_Family=mir-448|Gene=OR4F5
mature=hsa-miR-659-3p|mir_Family=-|Gene=OR4F5
mature=hsa-miR-5197-3p|mir_Family=-|Gene=OR4F5
mature=hsa-miR-5093|mir_Family=-|Gene=OR4F5
mature=hsa-miR-650|mir_Family=mir-650|Gene=OR4F5
df <- read.table(sep = "|", text = Data, stringsAsFactors = FALSE)
l <- lapply(df, strsplit, "=")
trim <- function(x) gsub("^\\s*|\\s*$", "", x)
paste(trim(sapply(l[[1]], "[", 2)), trim(sapply(l[[3]], "[", 2)), sep = "-")
# [1] "hsa-miR-5087-OR4F5" "hsa-miR-26a-1-3p-OR4F9" "hsa-miR-448-OR4F5" "hsa-miR-659-3p-OR4F5" "hsa-miR-5197-3p-OR4F5" "hsa-miR-5093-OR4F5"
# [7] "hsa-miR-650-OR4F5"
答案 4 :(得分:4)
也许不是更优雅,但你可以尝试:
sapply(Data[,1],function(x){
parts<-strsplit(x,"\\|")[[1]]
y<-paste(gsub("(mature=)|(Gene=)","",parts[grepl("mature|Gene",parts)]),collapse="-")
return(y)
})
示例
Data<-data.frame(col1=c("mature=hsa-miR-5087|mir_Family=-|Gene=OR4F5","mature=hsa-miR-26a-1-3p|mir_Family=mir-26|Gene=OR4F9"),col2=1:2,stringsAsFactors=F)
> Data[,1]
[1] "mature=hsa-miR-5087|mir_Family=-|Gene=OR4F5" "mature=hsa-miR-26a-1-3p|mir_Family=mir-26|Gene=OR4F9"
> sapply(Data[,1],function(x){
+ parts<-strsplit(x,"\\|")[[1]]
+ y<-paste(gsub("(mature=)|(Gene=)","",parts[grepl("mature|Gene",parts)]),collapse="-")
+ return(y)
+ })
mature=hsa-miR-5087|mir_Family=-|Gene=OR4F5 mature=hsa-miR-26a-1-3p|mir_Family=mir-26|Gene=OR4F9
"hsa-miR-5087-OR4F5" "hsa-miR-26a-1-3p-OR4F9"
相关问题
最新问题
- 我写了这段代码,但我无法理解我的错误
- 我无法从一个代码实例的列表中删除 None 值,但我可以在另一个实例中。为什么它适用于一个细分市场而不适用于另一个细分市场?
- 是否有可能使 loadstring 不可能等于打印?卢阿
- java中的random.expovariate()
- Appscript 通过会议在 Google 日历中发送电子邮件和创建活动
- 为什么我的 Onclick 箭头功能在 React 中不起作用?
- 在此代码中是否有使用“this”的替代方法?
- 在 SQL Server 和 PostgreSQL 上查询,我如何从第一个表获得第二个表的可视化
- 每千个数字得到
- 更新了城市边界 KML 文件的来源?